Archives
Among the GWAS loci with more frequent lead
Among the GWAS loci with more frequent lead SNPs (estimated MAF>5%) and corresponding validation GWAS or exome sequencing association signals, we observed p=3.34×10 for an intergenic SNP (rs9529561) on chromosome 13q21.33 which was also supported by the PROGRESS data (uncorrected p=0.035 for the association to death within 28days or necessity of intensive care; p=2.4×10 in the meta-analysis). While again no data was available from the meta-analysis by Rautanen et al. (2015), the exome data for the KLHL1 (kelch like family member 1) gene results in 375kb distance did not provide evidence for an association (pSKAT-O=1.00; Table 4). For an intronic SNP (rs2641697) of the CRISPLD2 (cysteine rich secretory protein LCCL domain containing 2) gene on chromosome 16q24.1, we observed p=5.99×10 in the discovery GWAS and pSKAT-O=0.003 for 28day mortality in the exome data. However, this time none of the independent validation GWA studies supported this observation. GTEx and ENCODE annotations for the two loci are provided in Supplementary Figs. 20 and 21.
Finally, we compared the top association findings (p≤10) from Rautanen et al. (2015) with the results from our discovery GWAS and exome sequencing study (Supplementary Table 2). Within the reported 12 loci with variants associated with 28day mortality among patients with sepsis caused by pneumonia or abdominal infections, we observed p=0.01 for rs2096460 located in URB1 (URB1 ribosome biogenesis 1 homolog (S. cerevisiae)) on chromosome 21q22.11 and p=0.04 for an intergenic variant (rs74438932) on chromosome 13 located 88kb downstream of GPR12 (G protein-coupled receptor 12) gene on chromosome 13q12.13. However, the effect casin in both GWAS were directionally inconsistent and none of the available exome sequencing association signals achieved a pSKAT-O≤0.05. For details we refer to Supplementary Table 2.
Discussion
We report results of a GWAS in patients wi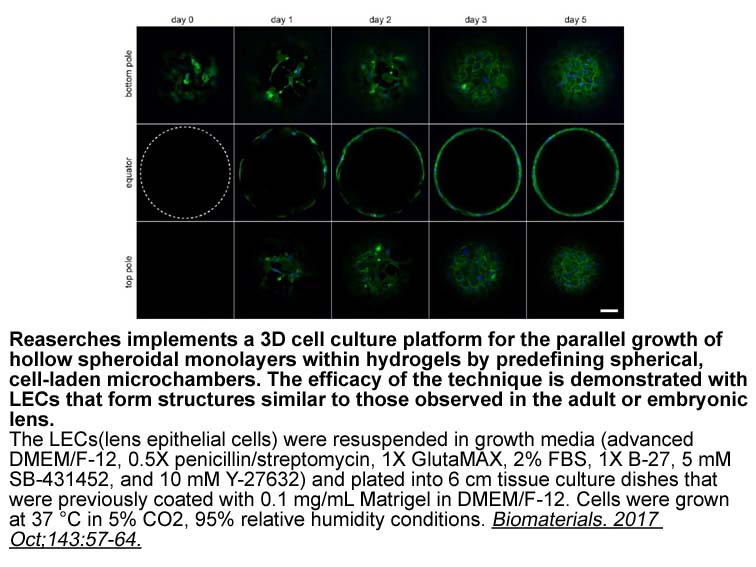 th treated sepsis which focused on common genetic variants associated with 28day mortality. We validated our best findings using three independent data sets including two GWA and a whole-exome sequencing study. We applied the new Sepsis-3 definition (Singer et al., 2016; Seymour et al., 2016) in the discovery GWAS and the sequencing study which requires the presence of organ dysfunction for a diagnosis of sepsis. Furthermore, we followed-up the best discovery loci from the most recent and largest sepsis GWAS which applied a similar study design (Rautanen et al., 2015).
The SNP with the strongest GWAS signal was a missense and potentially deleterious variant located on chromosome 9q21.2 within VPS13A. This result was supported by the validation GWAS and the exome sequencing data. Recent experiments (Muñoz-Braceras et al., 2015) indicated an important regulatory role of VPS13A for autophagic degradation. Autophagy is a key component of our immune system and has also been associated to several human diseases (Cuervo and Macian, 2014; Schneider and Cuervo, 2014). However, this signal is located within a gene-rich region (9q21) that has been associated to many complex diseases like mental disorders, type 2 diabetes mellitus, some cancers and cardiovascular disease (An et al., 2012; Shimo et al., 2011). Thus, VPS13A might not be the only candidate. Notably, we also observed a similarly strong association GWAS signal for variants in GNA14 (guanine nucleotide binding protein (G protein), alpha 14), a gene which is located ~200kb distal to VPS13A. GNA14 is member of the “G alpha Q signaling events”-pathway which is highlighted in the accompanying report by Taudien et al. 2016. In their report, rare variants in genes of the pathway, including GNA14, were found to have a putatively protective effect leading to a more favorable sepsis course. Identification of GNA14 in both studies is a strong argument that the G alpha Q signaling pathway might play an important role in sepsis. In contrast to our findings, Rautanen et al. (2015) did not list this 9q21 locus around VPS13A and GNA14 among their top findings. Potential explanations, apart from a false positive finding in our data sets, could be their exclusion of variants with minor allele frequencies <2% and/or inclusion of less severely affected patients by applying the 2001 sepsis definition (Levy et al., 2003) or the general need for much larger samples size as also underlined by our
th treated sepsis which focused on common genetic variants associated with 28day mortality. We validated our best findings using three independent data sets including two GWA and a whole-exome sequencing study. We applied the new Sepsis-3 definition (Singer et al., 2016; Seymour et al., 2016) in the discovery GWAS and the sequencing study which requires the presence of organ dysfunction for a diagnosis of sepsis. Furthermore, we followed-up the best discovery loci from the most recent and largest sepsis GWAS which applied a similar study design (Rautanen et al., 2015).
The SNP with the strongest GWAS signal was a missense and potentially deleterious variant located on chromosome 9q21.2 within VPS13A. This result was supported by the validation GWAS and the exome sequencing data. Recent experiments (Muñoz-Braceras et al., 2015) indicated an important regulatory role of VPS13A for autophagic degradation. Autophagy is a key component of our immune system and has also been associated to several human diseases (Cuervo and Macian, 2014; Schneider and Cuervo, 2014). However, this signal is located within a gene-rich region (9q21) that has been associated to many complex diseases like mental disorders, type 2 diabetes mellitus, some cancers and cardiovascular disease (An et al., 2012; Shimo et al., 2011). Thus, VPS13A might not be the only candidate. Notably, we also observed a similarly strong association GWAS signal for variants in GNA14 (guanine nucleotide binding protein (G protein), alpha 14), a gene which is located ~200kb distal to VPS13A. GNA14 is member of the “G alpha Q signaling events”-pathway which is highlighted in the accompanying report by Taudien et al. 2016. In their report, rare variants in genes of the pathway, including GNA14, were found to have a putatively protective effect leading to a more favorable sepsis course. Identification of GNA14 in both studies is a strong argument that the G alpha Q signaling pathway might play an important role in sepsis. In contrast to our findings, Rautanen et al. (2015) did not list this 9q21 locus around VPS13A and GNA14 among their top findings. Potential explanations, apart from a false positive finding in our data sets, could be their exclusion of variants with minor allele frequencies <2% and/or inclusion of less severely affected patients by applying the 2001 sepsis definition (Levy et al., 2003) or the general need for much larger samples size as also underlined by our  power considerations.
power considerations.